Solutions#
1. Sampling uniformly on a disk#
Consider the following probability distribution:
where \(\mathbb{1}_{x^2 + y^2 \leq 1}\) is the indicator function of the disk of radius 1 centered at the origin.
Using a transformation method, develop a sampler for this distribution.
Hint: Consider sampling a radius and an angle - and convert them to Cartesian coordinates. The most obvious choice may not work!
After the transformation method, implement the usual rejection sampling method to sample from the disk.
Sample from \(x\)-marginal of this distribution. Plot the histogram of the samples.
Solution: The naive approach is the sampler:
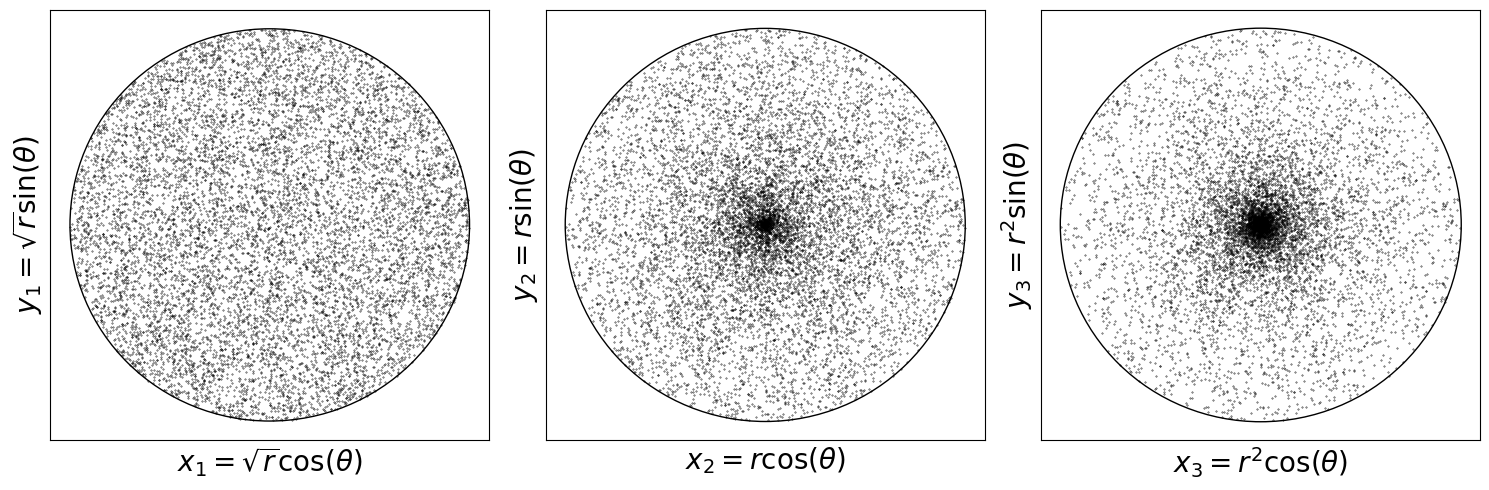
\(r \sim \text{Uniform}(0, 1)\) and \(\theta \sim \text{Uniform}(-\pi, \pi)\), then \(x = r \cos(\theta)\) and \(y = r \sin(\theta)\). This sampler is not uniform on the disk.
We can compute the density of \(X_1, X_2\) as
where \(g^{-1}\) is the inverse transformation. Let us construct the inverse transform:
by just observing that \(\cos^2 \theta + \sin^2 \theta = 1\). We can also write that
Therefore, we can write the inverse transformation as
We next need to compute the Jacobian matrix as:
Therefore, the determinant is
Therefore, the density of \(X_1, X_2\) is
Not uniform!
Correct answer is \(X_1 = \sqrt{r} \cos(\theta)\) and \(X_2 = \sqrt{r} \sin(\theta)\).
import numpy as np
import matplotlib.pyplot as plt
# sample an angle uniformly (0, 2pi)
def sample_angle(n):
return np.random.uniform(0, 2 * np.pi, n)
# sample a radius uniformly (0, 1)
def sample_radius(r_t, n):
return np.random.uniform(0, r_t, n)
n = 10000
r_t = 1
# sample n angles and n radii uniformly and plot them in cartesian coordinates
theta = sample_angle(n)
r = sample_radius(1, n)
x1 = np.sqrt(r) * np.cos(theta)
y1 = np.sqrt(r) * np.sin(theta)
x2 = r * np.cos(theta)
y2 = r * np.sin(theta)
x3 = r**2 * np.cos(theta)
y3 = r**2 * np.sin(theta)
# draw a unit circle and plot x, y samples within it
fig, ax = plt.subplots(1, 3, figsize=(15, 5))
ax[0].add_artist(plt.Circle((0, 0), np.sqrt(r_t), color='k', fill=False))
ax[0].scatter(x1, y1, s=0.1, c='k')
ax[0].set_xlabel(r"$x_1 = \sqrt{r} \cos(\theta)$", fontsize=20)
ax[0].set_ylabel(r"$y_1 = \sqrt{r} \sin(\theta)$", fontsize=20)
ax[0].set_xticks([])
ax[0].set_yticks([])
ax[1].add_artist(plt.Circle((0, 0), r_t, color='k', fill=False))
ax[1].scatter(x2, y2, s=0.1, c='k')
ax[1].set_xlabel(r"$x_2 = r \cos(\theta)$", fontsize=20)
ax[1].set_ylabel(r"$y_2 = r \sin(\theta)$", fontsize=20)
ax[1].set_xticks([])
ax[1].set_yticks([])
ax[2].add_artist(plt.Circle((0, 0), r_t, color='k', fill=False))
ax[2].scatter(x3, y3, s=0.1, c='k')
ax[2].set_xlabel(r"$x_3 = r^2 \cos(\theta)$", fontsize=20)
ax[2].set_ylabel(r"$y_3 = r^2 \sin(\theta)$", fontsize=20)
ax[2].set_xticks([])
ax[2].set_yticks([])
fig.tight_layout()
plt.show()
2. Monte Carlo Integration#
Consider the following integral:
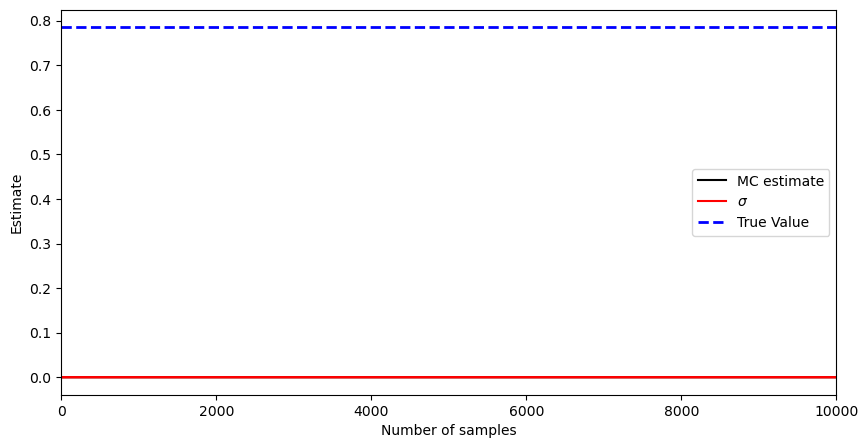
Take the sampling distribution as uniform on \([0,1]\). Build the Monte Carlo estimate for varying \(N\) and compute the mean and the variance, i.e., for \(\varphi(x) = (1 - x^2)^{1/2}\) and given \(X_1, \ldots, X_N\) from \(\text{Unif}(0,1)\), compute
and the empirical variance
Discuss the difference between this empirical estimator vs. the correct one for the variance (using the true value). Plot your mean estimates \eqref{eq:mean_estimates}, standard deviation estimates (by taking the square root of \eqref{eq:variance_est}), and the true value \(I = \pi/4\). Can you always trust the variance estimates? Here is a code snippet for this one:
import numpy as np
import matplotlib.pyplot as plt
def phi(x):
return np.sqrt((1 - x**2))
I = np.pi / 4 # true value
N_max = 10000 # go up to 10,000 samples
U = np.random.uniform(0, 1, N_max)
I_est = np.zeros(N_max - 1) # this is longer than we need
I_var = np.zeros(N_max - 1)
fig = plt.figure(figsize=(10, 5))
k = 0
K = np.array([])
# We are not computing for every N for efficiency
for N in range(1, N_max, 5):
I_est[k] = 0 # Your mean estimate here
I_var[k] = 0 # Your variance estimate here
k = k + 1 # We index estimators with k as we jump N by 5
K = np.append(K, N)
plt.plot(K, I_est[0:k], 'k-', label='MC estimate')
plt.plot(K, I_est[0:k] + np.sqrt(I_var[0:k]), 'r', label=r'$\sigma$', alpha=1)
plt.plot(K, I_est[0:k] - np.sqrt(I_var[0:k]), 'r', alpha=1)
plt.plot([0, N_max], [I, I], 'b--', label='True Value', alpha=1, linewidth=2)
plt.legend()
plt.xlabel('Number of samples')
plt.ylabel('Estimate')
plt.xlim([0, N_max])
(0.0, 10000.0)
Solution:
import numpy as np
import matplotlib.pyplot as plt
def phi(x):
return np.sqrt((1 - x**2))
I = np.pi / 4
N_max = 100000
U = np.random.uniform(0, 1, N_max)
I_est = np.zeros(N_max - 1)
I_var = np.zeros(N_max - 1)
I_var_correct = np.zeros(N_max - 1)
fig = plt.figure(figsize=(10, 5))
K = np.array([])
k = 0
for N in range(1, N_max, 1):
# print(N)
I_est[k] = (1/N) * np.sum(phi(U[0:N]))
I_var[k] = (1/(N**2)) * np.sum((phi(U[0:N]) - I_est[k])**2)
k = k + 1
K = np.append(K, N)
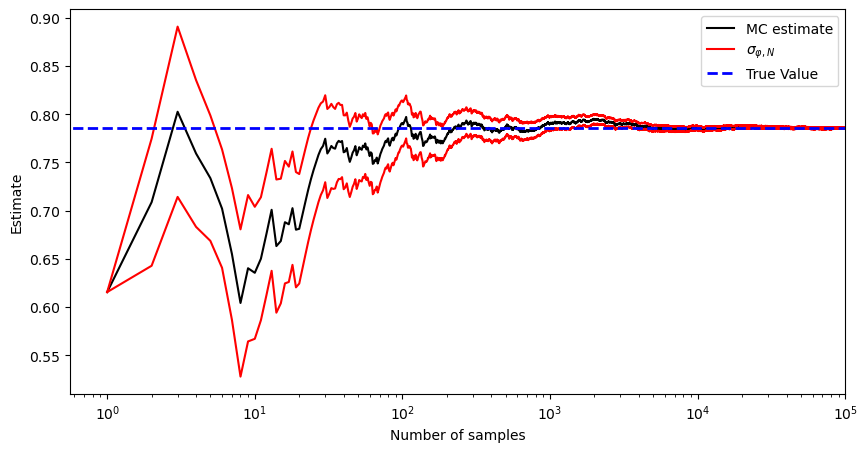
plt.semilogx(K, I_est[0:k], 'k-', label='MC estimate')
plt.plot(K, I_est[0:k] + np.sqrt(I_var[0:k]), 'r', label='${\\sigma}_{\\varphi,N}$', alpha=1)
plt.plot(K, I_est[0:k] - np.sqrt(I_var[0:k]), 'r', alpha=1)
plt.plot([0, N_max], [I, I], 'b--', label='True Value', alpha=1, linewidth=2)
plt.legend()
plt.xlabel('Number of samples')
plt.ylabel('Estimate')
plt.xlim([0, N_max])
plt.show()
/var/folders/d8/86whr2t12h70n511w7s7cm_w0000gn/T/ipykernel_14736/2275840071.py:38: UserWarning: Attempt to set non-positive xlim on a log-scaled axis will be ignored.
plt.xlim([0, N_max])
3. Importance Sampling#
Implement the importance sampling method for estimating
where \(X\sim \mathcal{N}(0,1)\). Try two methods: (i) Standard Monte Carlo integration by drawing i.i.d samples from \(\mathbb{P}\) and (ii) importance sampling. What kind of proposals should you choose? What is a good criterion for this example? Choose different proposals and test their efficiency in terms of getting a low relative error vs. samples.
Solution: Monte Carlo sampler is just sampling \(\mathcal{N}(0,1)\) and keeping samples \(X>4\) as we covered several times – the IS estimator is also provided in lecture notes. Below is one solution. Here the proposal is $\(q(x) = \mathcal{N}(x; 6, 1).\)$
import numpy as np
import matplotlib.pyplot as plt
xx = np.linspace(4, 20, 100000)
def p(x):
return np.exp(-x**2/2)/np.sqrt(2*np.pi)
def q(x, mu, sigma):
return np.exp(-(x-mu)**2/(2*sigma**2))/(np.sqrt(2*np.pi)*sigma)
def w(x, mu, sigma):
return p(x)/q(x, mu, sigma)
I = np.trapezoid(p(xx), xx) # Numerical computation of the integral
print('Integral of p(x) from 4 to infinity: ', I)
N = 10000
x = np.random.normal(0, 1, N) # iid samples from p(x)
I_est_MC = (1/N) * np.sum(x > 4)
print('Monte Carlo estimate: ', I_est_MC)
mu = 6
sigma = 1
x_s = np.zeros(N)
weights = np.zeros(N)
for i in range(N):
x_s[i] = np.random.normal(mu, sigma, 1)[0]
weights[i] = w(x_s[i], mu, sigma)
I_est_IS = (1/N) * np.sum(weights * (x_s > 4))
print('Importance sampling estimate: ', I_est_IS)
Integral of p(x) from 4 to infinity: 3.16712429751607e-05
Monte Carlo estimate: 0.0
Importance sampling estimate: 3.0485644265385348e-05
